 |
 |
|
|
|||
 |
|||
Trinucleotide (TRIMER CODON) Phosphoramidites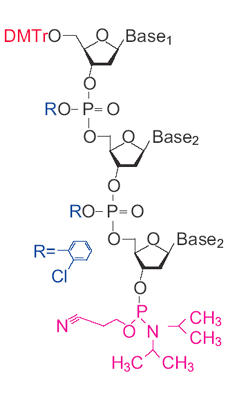 Catalogue number: 103-20 Description: white to off-white powder Storage of dry compound: 1 year at -20ºC Protein mutagenesis can be used to fine tune a variety of properties, such as improved stability to high temperatures, denaturants, or non- aqueous solvents; higher affinity binding to a target molecule; increased rates of enzymatic reactions; or changes of specificities. However, generating and finding these improved proteins can be a difficult task. One of the most popular methods is to make pools of degenerate oligonucleotides, which can be incorporated into the genes as cassettes or by PCR by using the degenerate oligo as a primer.1 Degenerate oligonucleotides are synthesized as a mixture of A/C/G/T phosphoramidites (N) at the site of the codons to be mutated. Problems arise, though, from using an equimolar solution of each base. First there is a coding bias. Out of the 64 possible codon combinations of A, C, G and T, 18 code for leucine, arginine or serine, but only 2 for tryptophan or methionine. As a result, only 3% of the mutagenic oligonucleotides will contain methionine or tryptophan, and over 28% will contain either leucine, arginine or serine. In addition, the three nonsense codons will lead to chain termination in 4.7% of the sequences. There are ways to improve this situation. For instance, using two degenerate mixes of bases, N and G/C, on the DNA synthesizer to insert NNG/C into the sequence will halve the number of the most degenerate codons, but still code for all 20 amino acids. However, still 59% of the clones will code for just eight amino acids and 3% will have a stop codon inserted. The generation of redundant sequences and stop codons makes searching a clonal library inefficient. However, it is possible to improve the efficiency of this process by using a mixture of trinucleotide (trimer) phosphoramidites.2–5 By synthesizing a set of trimers that cover all 20 amino acids, the mutation of a gene can be carried out at the codon level rather than at individual bases. Therefore, unlike other methods of mutagenesis, trimer phosphoramidites lead to no codon bias, no frame-shift mutations, and no production of stop codons, making them one of the most efficient tools to explore sequence space in protein regions that are important for function 6 – even in nonsaturating conditions.7, 8 References: 1. Zon, G., Gallo, K., Samson, C., Shao, K., Michael F. Summers, M., Byrd, R. Nucleic Acids Res, 1985, 13, 8181-8196. 2. Kayushin, A., Korosteleva, M., Miroshnikov. Nucleos. Nucleot. Nucleic Acids, 2000, 19, 1967-1976. 3. Kayushin, A., M. Korosteleva, . Miroshnikov, A. Nucleos Nucleot, 1999, 18, 1531-1533. 4. Kayushin, A., M. Korosteleva, . Miroshnikov, A. W. Kosch, W., Zubov, D., Piel N. Nucleic Acids Res., 1996, 24, 3748-3755. 5. Mauriala, T., Auriola, S., Azhayev, A., Kayushin, A., Korosteleva, M., Miroshnikov, A. J Pharm Biomed Anal, 2004. 34, 199-206. 6. Yagodkin, A., Azhayev, A., Roivainen, J., Antopolsky, M., Kayushin, A., Korosteleva, M., Miroshnikov, A., Randolph, J., Mackie, H. Nucleos. Nucleot. Nucleic Acids 2007, 26, 473-497. 7. Neylon, C. Nucleic Acids Res, 2004. 32, 1448-59. 8. Sondek, J. and D. Shortle, Proc Natl. Acad. Sci. U S A, 1992. 89,3581-3585. Price and Ordering Information |
|||